Worm plots are back in UCSF ChimeraX
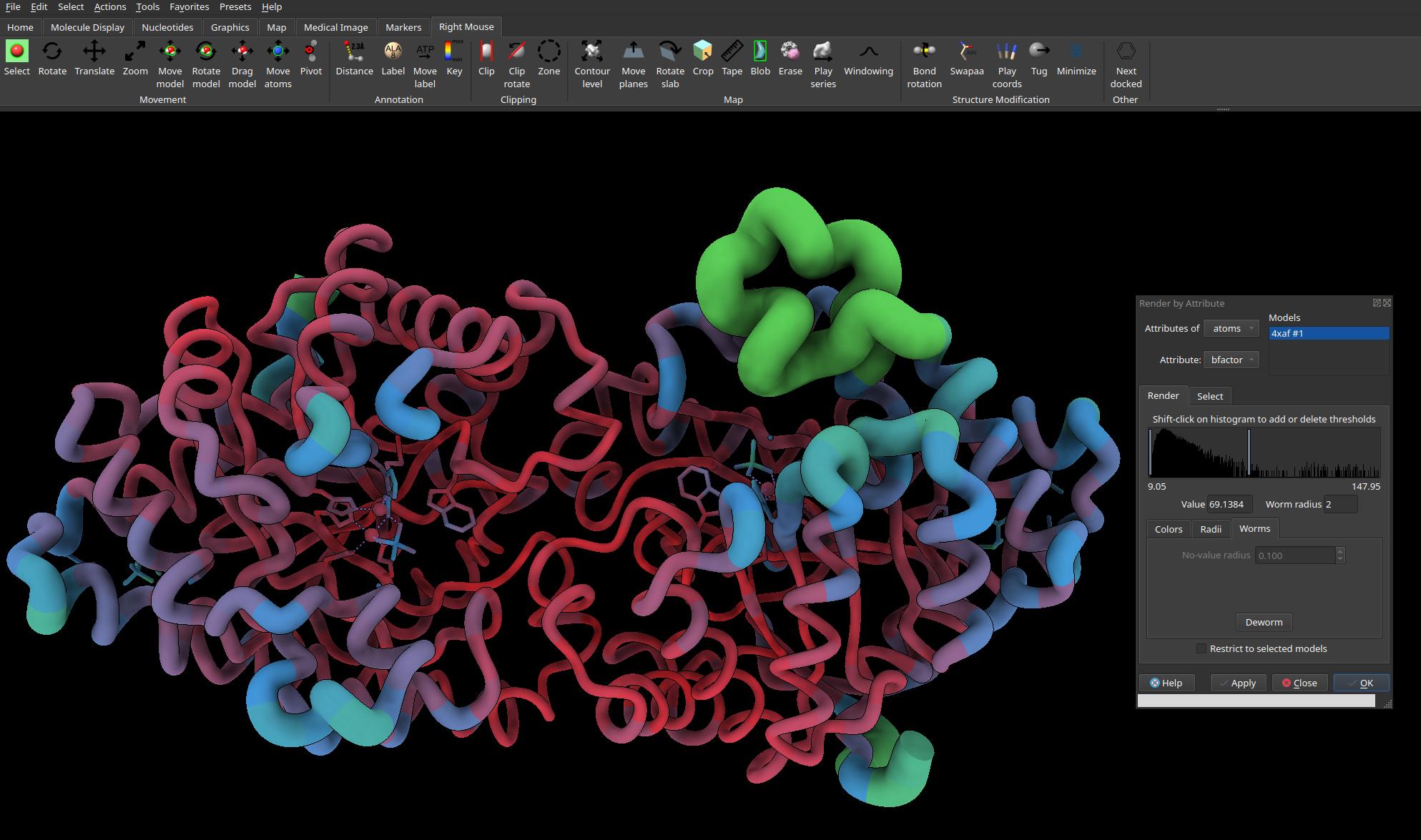 B-factors of crystal structure represented by “worms” (PDB: 4XAF)
B-factors of crystal structure represented by “worms” (PDB: 4XAF)
Finally, as of the 13th March 2024, worm plots are back in the latest daily build version of UCSF ChimeraX (v1.8). Worm plots used to be a feature of old Chimera (which is still available on the website archive), but was removed from its successor ChimeraX. That was a shame because I thought worm plots look so visually stunning. Aside from being visually stunning, they’re also a useful way to depict B-factor values in crystallography structures, and B-factors are a useful proxy for inferring conformational dynamics.1
Below are a couple of examples I’ve seen in the literature of worm plots used to depict B-factors and conformational dynamics:
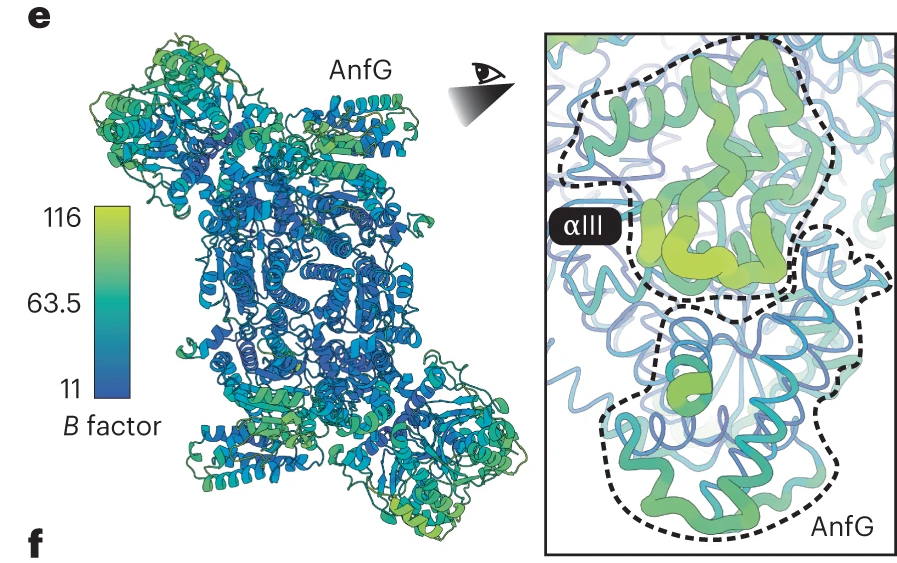 Schmidt et al. (2024)2
Schmidt et al. (2024)2
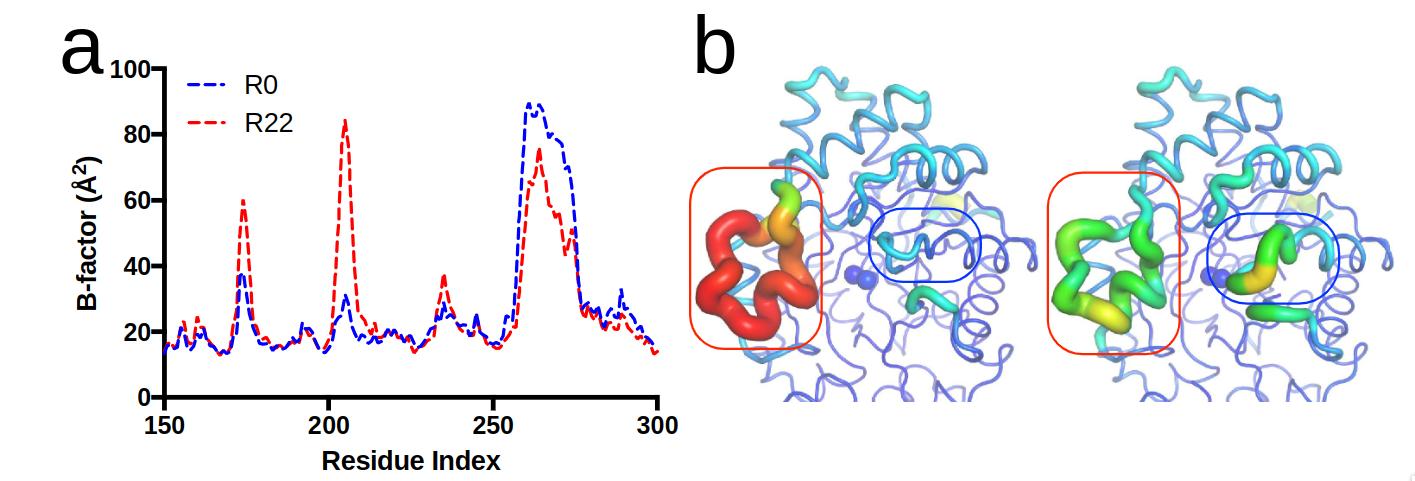 Campbell et al. (2016)3
Campbell et al. (2016)3
How to make worm plots in ChimeraX
In your daily build version of ChimeraX, simply go to:
- Tools $\rightarrow$ Depiction $\rightarrow$ Render by Attribute
- In the “Render by Attribute” pop-up window, set:
- “Attributes of” to atoms
- “Attribute” to bfactor
- Then in the “Worms” tab below, adjust the size of the worms according to your needs
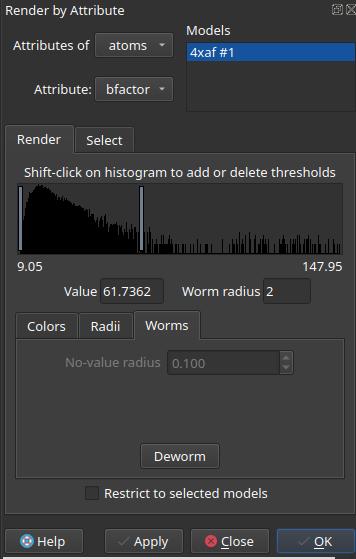
For more options, see the “Cartoon by Attribute (Worm)” help page in the ChimeraX built-in help docs (help:user/commands/cartoon.html#byattribute) or the online docs for the same page. The syntax of the command are as follow:
- Set worm plot with specific parameters:
- ( cartoon byattribute | worm ) attribute-name model-spec [ values-and-radii ] [ noValueRadius radius ] [ sides N ]
- Set worm plot on or off:
- ( cartoon byattribute | worm ) ( on | off )
- Toggle worm plot on or off:
- ~worm
For example, cartoon byattr bfactor #1 min:0.2 max:2.5 is equivalent to worm bfactor #1 min:0.2 max:2.5.
The parameters noValueRadius and sides are best left at their default values by exclusing them from the command. I’ve tried messing around with them, but anything other than the default values only makes the worm plots doesn’t look nice (unless of course you like the look of cubic worms by setting sides to 4).
References
Li DW, Brüschweiler R. All-atom contact model for understanding protein dynamics from crystallographic B-factors. Biophys J. 2009 Apr 22;96(8):3074-81. doi: 10.1016/j.bpj.2009.01.011. PMID: 19383453; PMCID: PMC2718318. ↩︎
Schmidt, F.V., Schulz, L., Zarzycki, J. et al. Structural insights into the iron nitrogenase complex. Nat Struct Mol Biol 31, 150–158 (2024). https://doi.org/10.1038/s41594-023-01124-2 ↩︎
Campbell, E., Kaltenbach, M., Correy, G. et al. The role of protein dynamics in the evolution of new enzyme function. Nat Chem Biol 12, 944–950 (2016). https://doi.org/10.1038/nchembio.2175 ↩︎