Adding cofactors to AlphaFold2 models with help from FoldSeek
Table of Contents
AlphaFold2 can’t predict cofactors (yet)
One of the limitations of AlphaFold2 is that it can’t predict where cofactors or ligands will bind to in a protein. It does however, do an eerily good job of predicting the structural architecture of cofactor/ligand binding sites. For instance, pictured below is a hydrogenase structure I’m currently investigating. Its structure was predicted with AF2 (ColabFold, to be more precise). Hydrogenases contain iron-sulfur (FeS) clusters and the Fe atoms bond with the S atoms of cysteine residues. The AF2 predicted hydrogenase structure does a great job of predicting where the four cysteine residues would coordinate with a [4Fe-4S] cluster. You can see where the cluster should go into the structure. 1
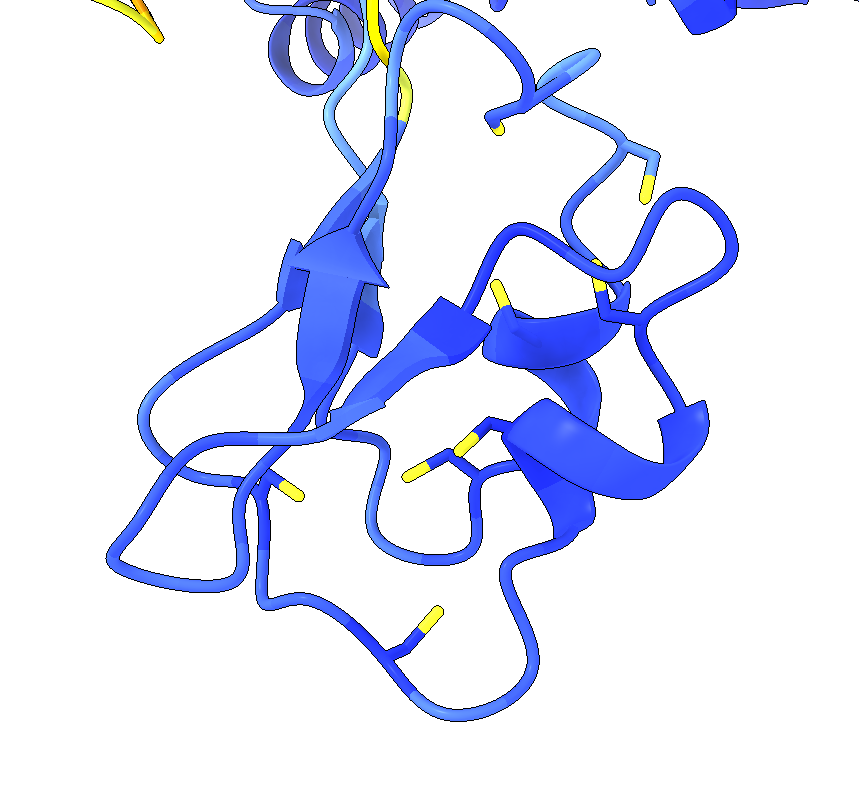 An “apo” hydrogenase AF2 model
An “apo” hydrogenase AF2 model
Such a structure is almost certainly biophysically impossible and would never exist in reality because the protein would collapse into the solvent void where the FeS cluster should be. At the same time this highlights the unique advantage of structural programs that don’t take biophysical principles into account, like AlphaFold2. Such a program is able to predict protein structure much more quickly and easily, and at some point other programs can “fill in” the missing cofactors/ligands if needed.
One such program is AlphaFill (alphafill.eu), which aims to “transplant” missing ligands, cofactors, and metal ions into “apo” AF2 models. The program works by searching the PDB for sequence homologs of your query model and then aligning/overlaying the structures. The confactors/ligands within the structures output from the PDB are then superimposed with the query model and given a Local RMSD score, and a Transplant Clash Score (TCS). Local RMSD is what it sound like; it’s the RMSD of all Cα’s within 6Å of the ligand. The TCS is the most relevant score, as that’s the RMSD of the van der Waals overlap between overlap and query model.
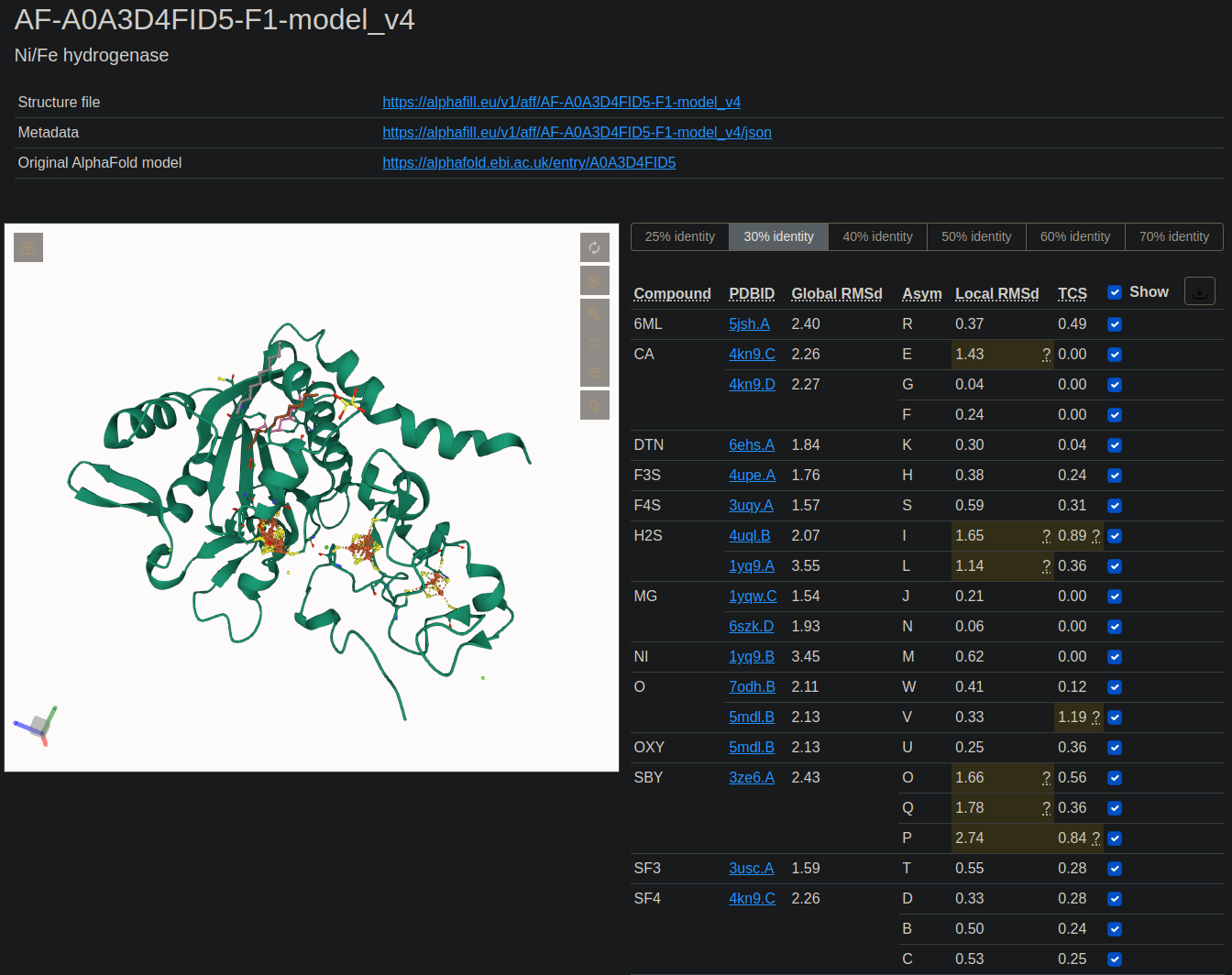 Example of the AlphaFill database results window
Example of the AlphaFill database results window
I’ve noticed some unfortunate limitations with the AlphaFill method in my own queries. For starters, it struggles with AF2 query models that have distant sequence homology to structures in the PDB. So even if the query model has high structural similarity to PDB structures, AlphaFill may not find them due to the lack of sequence homology. Relying on sequence homology, rather than structural similarity, tends to return unhelpful results.
FoldSeek to the rescue
I’ve found a workaround with the help of FoldSeek. FoldSeek is relatively new at the time of writing this, but it’s truly amazing. It’s a program for finding structural similarity between proteins, similar to programs like Dali and TM-align. However, unlike Dali and TM-align, FoldSeek is:
- Extraordinarily fast while still being accurate
- Searches more databases, including the AlphaFold model database
- Can be run with a command-line interface or on a beautiful and user-friendly webserver UI: foldseek.com
Even if two proteins with highly similar structures share little to no sequence homology, FoldSeek has no trouble finding them and aligning them. That’s because the FoldSeek method doesn’t rely on sequence at all; instead it encodes each C$\alpha$ and the neighbouring C$\alpha$’s in 3D space as a single piece of information that summarises all their 3-Dimensional co-ordinates. By stringing together short blocks of these encodings (essentially representing a short sequence of the protein), FoldSeek is able to quickly search structural databases in a similar fashion to sequence searching (at least, this is how I understand it).
My experience using FoldSeek has been very fun and I’ve used it to address the same problem that AlphaFill tries to
solve (albeit, in a slower, more manual fashion).
When I searched AlphaFill with my hydrogenase of interest (see above), the cofactor “transplants” it output were quite obviously erroneous.
There were lots of steric clashes and the output structures had very little structural similarity to my query.
However, using FoldSeek to search the “PDB100” database returned numerous highly similar structures that AlphaFill
missed (many of which are hydrogenases as well, which was a promising sign).
The FoldSeek web UI is also slick in that you can visualise which regions of the superimposed proteins are overlapping
or not.
On closer inspection of the PDBs on some of the results, I could pick PDBs that contained the relevant FeS clusters and
open those PDBs and my AF2 hydrogenase model of interest in UCSF ChimeraX.
Then using the matchmaker command to align the structures, I could see if any of the FeS clusters in the PDB
structures aligned with where I’d expect them to sit in my AF2 model.
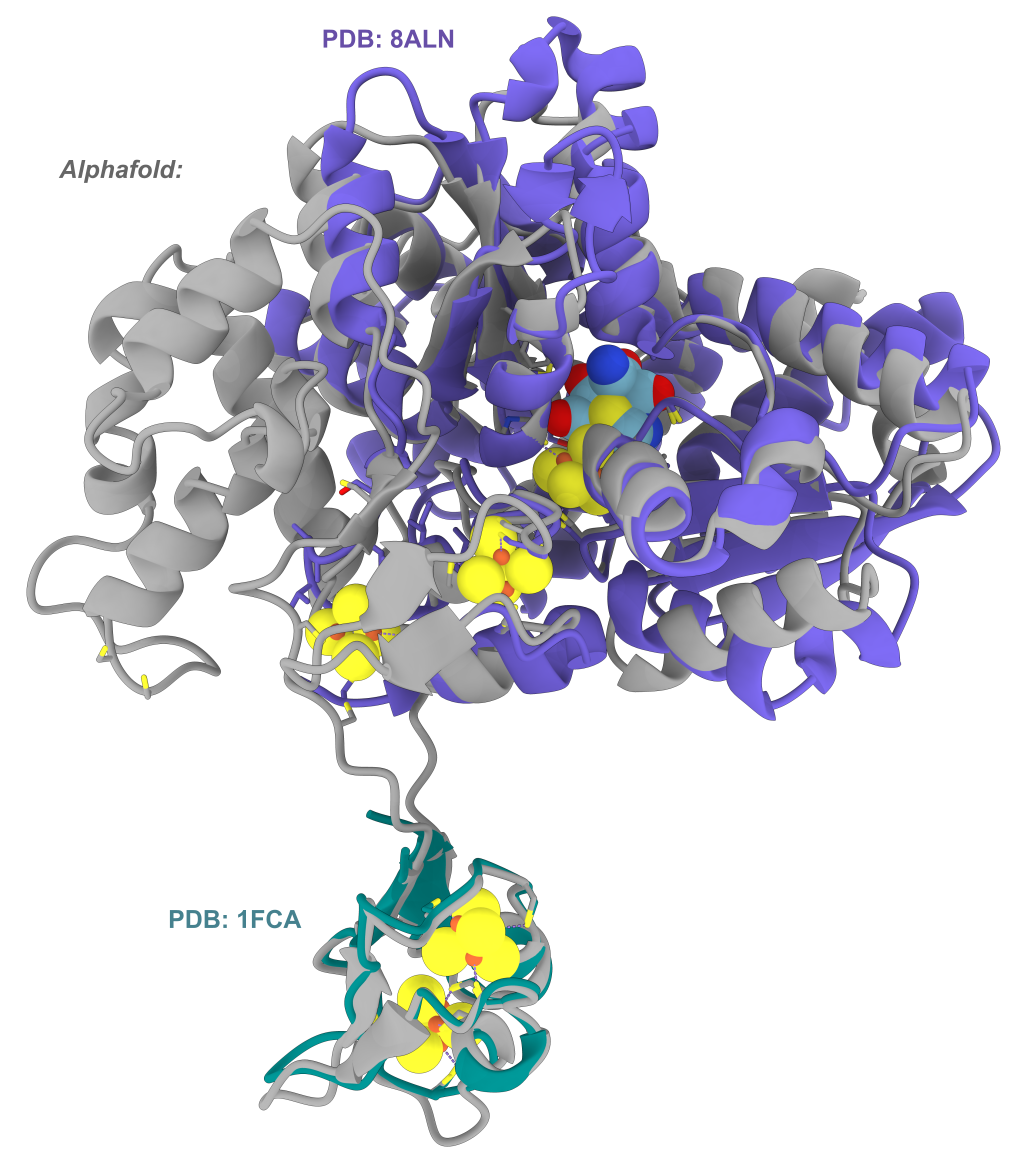 My AF2 model aligned with the two experimental structures found by FoldSeek
My AF2 model aligned with the two experimental structures found by FoldSeek
Taking a closer look at the FeS binding sites, we see an almost perfect overlap between my AF2 hydrogenase and the PDB hydrogenase FoldSeek found for me.
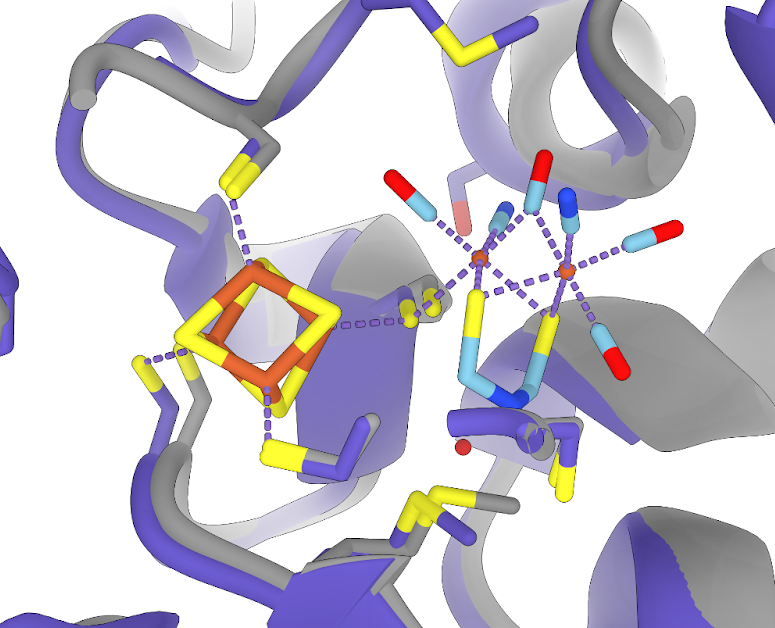 FeS clusters from the PDB line up well with the AF2 model
FeS clusters from the PDB line up well with the AF2 model
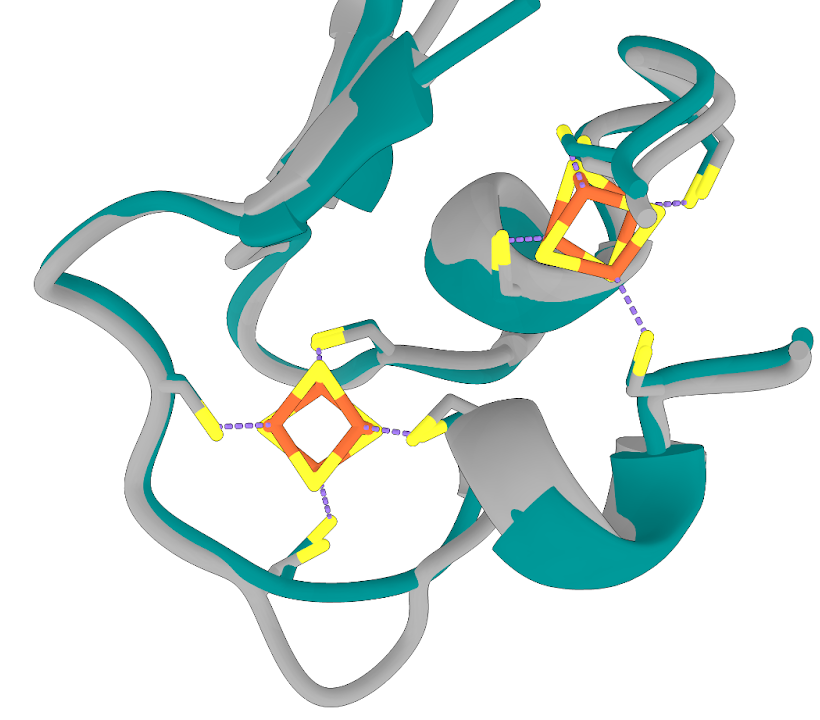 The structures here are so well aligned that “transplanting” the FeS clusters requires no manual readjustment in
ChimeraX
The structures here are so well aligned that “transplanting” the FeS clusters requires no manual readjustment in
ChimeraX
From there it was a simple matter of “transplanting” the cofactors from the PDBs into the AF2 model in ChimeraX. Only one of the cofactors required a fair deal of manual repositioning in space.
What the future holds
This approach to “filling” an AF2 model with its missing cofactors/ligands is fun, though admittedly more of an art than a science. For the time being, it’s a quick fix to this problem. However, it may not be a problem for much longer, as Google’s DeepMind and Isomorphic Labs announced that they’ve developed the next iteration of AlphaFold that can predict cofactors/ligands too: A Glimpse of the next Generation of AlphaFold